Whole transcriptome Spatial Services
Submit your tissue samples, and we’ll handle everything—from slide preparation and imaging to sequencing and spatial analysis. Receive high-resolution spatial gene expression maps with full data outputs and expert support.
Why perform Spatial Transcriptomics?
Spatial transcriptomics is a powerful research tool because it allows scientists to map gene expression within the precise anatomical context of a tissue. Unlike traditional RNA-seq or even single-cell RNA-seq, which lose spatial information during sample preparation, spatial transcriptomics preserves the location of each transcript. This technology helps researchers understand not just what genes are expressed, but where they are expressed, revealing critical insights into tissue organization, cell-to-cell interactions, disease microenvironments, and developmental processes. It’s particularly transformative in fields like cancer research, neuroscience, immunology, and regenerative medicine, where spatial relationships between cells are key to understanding function and pathology.
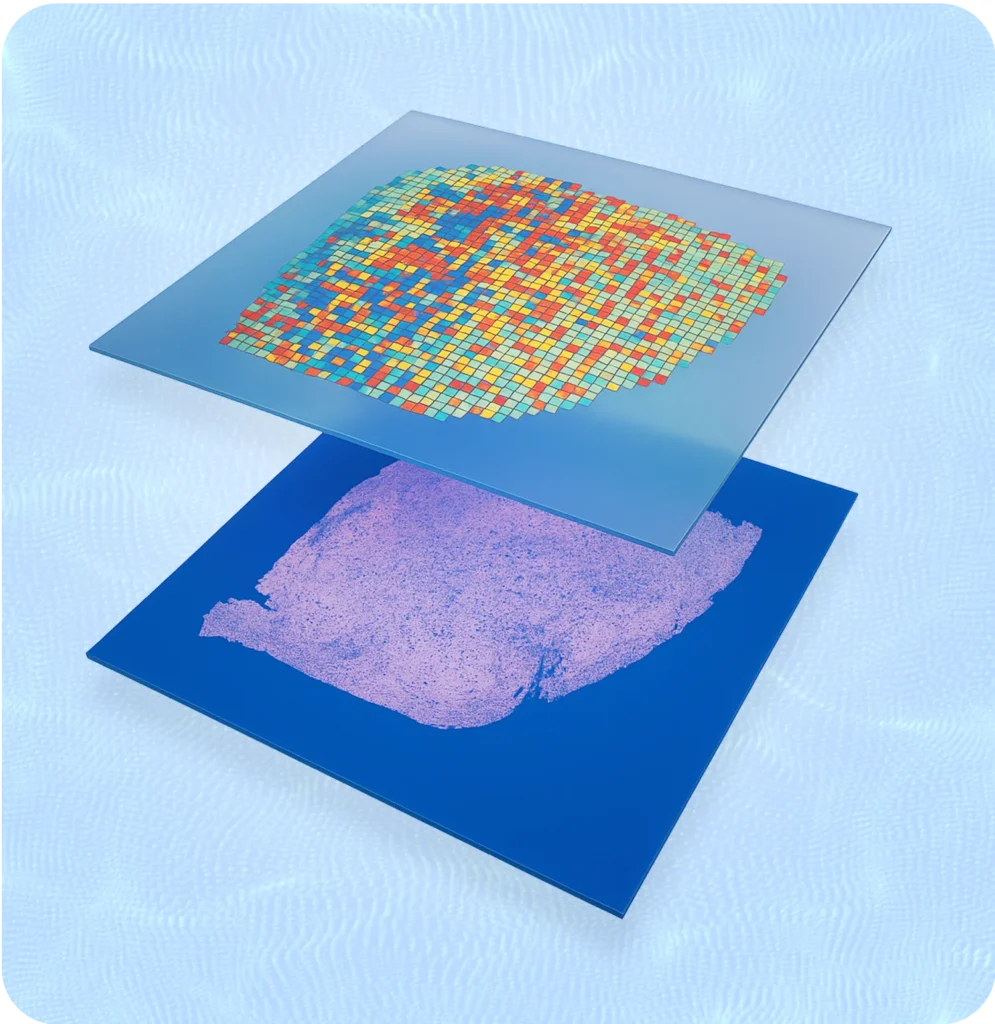

3D Genomics - Visium Expertise
At 3D Genomics our main expertise lies in the use of Visium and Visium HD, a whole-transcriptome spatial method developed by 10X Genomics.
10x Genomics’ Visium Spatial Gene Expression platform combines tissue imaging with RNA sequencing to map gene activity directly onto a tissue section. A fresh-frozen or FFPE tissue slice is placed on a specialized slide containing thousands of spots, each with unique barcoded oligonucleotides. As mRNA from the tissue binds to these barcodes, it captures spatial information linked to gene expression. After sequencing, the data is aligned with the tissue image, creating a detailed map showing where specific genes are active across the sample.
10x Genomics
Visium HD

Preserves spatial context of gene expression within the tissue architecture.
High-throughput — captures thousands of transcripts across large tissue sections.
Compatible with fresh-frozen and FFPE samples, expanding sample options.
Integrates imaging and sequencing for a more complete biological picture.
Enables discovery of cell niches, tissue organization, and disease microenvironments.
Flexible analysis — can be combined with histology, immunofluorescence, and other omics data.
Spatial transcriptomics + single-cell RNA-seq: a powerful combination
You can combine spatial and single-cell data by first using spatial technologies to map where gene expression occurs across a tissue and then integrating that with single-cell RNA-seq data to resolve the identities and states of individual cells within that spatial map. Single-cell profiles provide high-resolution gene expression signatures, while spatial data adds location context — together offering a more complete, detailed view of tissue architecture, cell types, and interactions.

Our Process
What you provide
- FFPE Samples: Blocks or on Slides (Human or Mouse)
- Frozen Tissue: We can fix or embed for use in Visium or other spatial technologies compatible with fresh frozen tissue (can work with most species).
What 3D Genomics provides
- General histological services: Tissue Fixation, H&E staining, Tissue Sectioning, CytAssist transfer (if applicable)
- Spatial Library preparation (including QC for sequencing and sequencing at depth determined during consultation).
- Sequencing FASTQ, read mapping and alignment using Space Ranger or other informatic pipelines and tools.
- Bioinformatics on mapped reads including clustering, cell type annotation and differential gene expression

What 3D Genomics delivers
- Raw sequencing files (FASTQ) and sequencing QC report.
- For 10x Assays the 10x Space Ranger output: web summary, feature-barcode matrices, BAM files, automated secondary analyses, Loupe browser files and other output files
- Computational support including clustering, cell type annotation and multiomic integration, if applicable
- Remaining tissue samples will be discarded or returned, as per your instructions
Ready to move your spatial and single cell research forward?
Partner with 3D Genomics for high-resolution spatial and single-cell transcriptomics services that move your research forward.