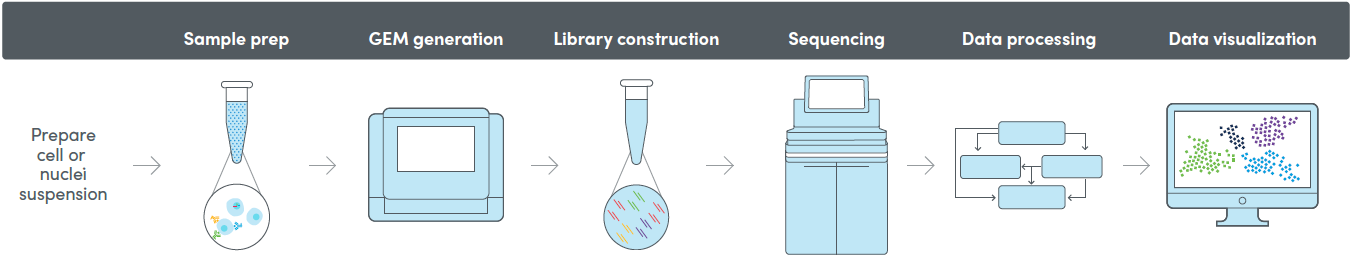
10X single-cell / single-nuclei RNA-seq
Single-cell RNA sequencing is a powerful tool for characterizing the whole transcriptome of cell populations. We provide end-to-end single-cell gene expression services, from cell preparation to bioinformatics analysis.
Single-cell or single-nuclei?
Your cell source and tissue type will determine whether gene expression libraries should be made from single-cell or single-nuclei suspensions.
Most cell culture samples can be effectively dissociated to single cells for scRNAseq and shipped to us as cryopreserved frozen cells.
Learn More
In many cases, fresh tissue can be dissociated to single cells. We use the Miltenyi GentleMACS Octo tissue dissociator for that purpose which can improve the yield and quality of dissociated cells. In general, frozen tissues are difficult to dissociate to single cells without significant cell loss or RNA degradation, whereas nuclei can be obtained from most fresh frozen tissues. For this reason we recommend single nuclei RNAseq from frozen tissue samples. While nuclei can be obtained from FFPE tissue blocks, a validated protocol for single-nuclei RNA sequencing (snRNA-seq) on the 10X Chromium platform has not yet been established.
Single-nuclei RNAseq detects fewer genes compared to single-cell RNAseq due to the lower mRNA content in nuclei, but this effect is modest and sample-dependent. Most studies show snRNA-seq captures gene expression largely comparable to scRNA-seq. At 3DG, we have performed successful nuclei isolation from multiple tissues.
Sample preparation
The quality of your single-cell suspension is the most important factor in acquiring high-quality sequencing libraries for single-cell gene expression. If cell viability is low, we can perform a dead cell removal kit to enrich for living cells. Viability above 90% is a good number for obtaining high quality sequencing libraries. If samples have a lower viability, dead cell removal can be used to improve accuracy in target cell count, increase library complexity, and decrease the percentage of mitochondrial genes in the final libraries.
Options for single-cell/single-nuclei RNA-seq
10X Genomics provides several options for generating the best gene expression libraries for your tissue. In addition to the standard whole-transcriptome approach, you may also consider additional single-cell options:
– Gene Expression with Feature Barcode technology – CRISPR screening, cell surface protein expression, and cell multiplexing
– Single-cell immune profiling
– Single-cell ATAC or multiome ATAC + gene expression to simultaneously measure gene expression and open chromatin
– Single-cell Gene Expression “LT” (low throughput) for small numbers of cells – great for pilot studies or rare cell populations
How to Work with Us
What you provide
Tissue samples or single-cell suspensions
- Single-cell suspensions should be at a minimum concentration of 400 cells/ul with a viability greater than 70%, with at least 50K cells total per sample.
What we provide
Cell preparation including dissociation, dead cell removal and/or nuclei isolation, if applicable
10X single-cell gene expression cDNA synthesis and library preparation
Sequencing, read mapping and alignment using CellRanger
Bioinformatics on mapped reads including clustering, cell type annotation and differential gene expression
What we deliver
Raw sequencing files (FASTQ) and sequencing QC report. 10X recommends sequencing a minimum of 20,000 read pairs per cell. However, in our experience most cells require 50,000+ read pairs per cell to reach a good saturation level. Sequencing depth will depend on cell type (high or low RNA content) and the goals of your experiment.
10X CellRanger output: web summary, feature-barcode matrices, BAM files, automated secondary analyses, Loupe browser files and other output files
Computational support including clustering, cell type annotation and multiomic integration, if applicable
Remaining tissue samples will be discarded or returned, as per your instructions